I apologize. I have made a comment in this link as well
Here is my cvx function that has to infer a valid Laplacian matrix
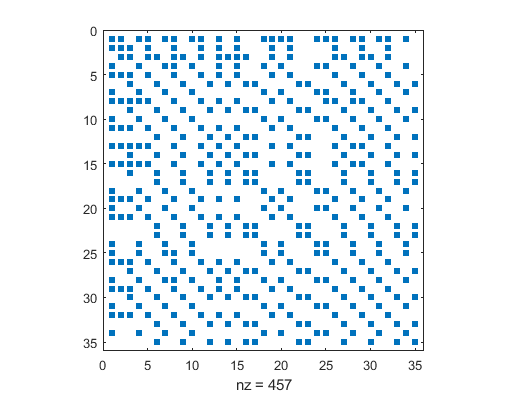
Here is my Laplacian’ spy plot/sparsity
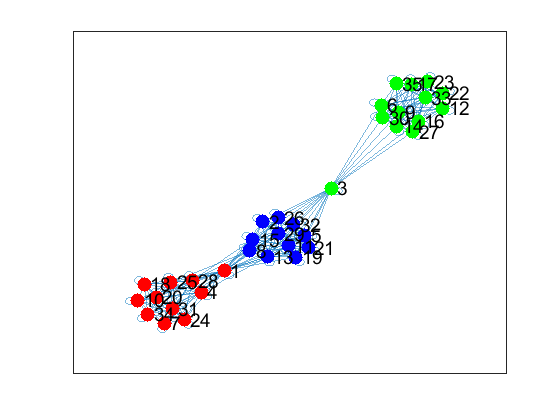
Here is the graph visualization from the inferred Laplacian
The constraints:
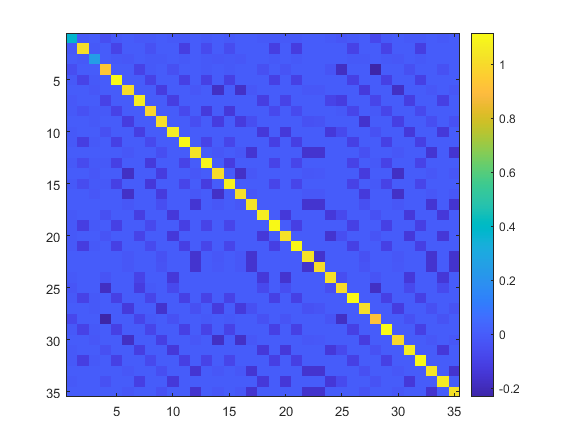
- issymmetric(L_D1norm) gives logical 1
- for positive semidefiniteness I am testing on the cholesky factorization
[~,p1]=chol(L_D1norm)
p1 =
0
Hence it looks like it has positive eigen values
3. min(eig(L_D1norm)<0) gives logical 0
4. trace(L_D1norm)=35 which are the number nodes on the graph
5. >> find(vec(L_D1norm-diag(diag(L_D1norm)))>0)
ans =
0×1 empty double column vector
which means that I have non-negative off diagonal entries
The graph visually gives a good partition into three distinct clusters which is what I needed.
This was what it was showing:
Calling SDPT3 4.0: 1898 variables, 631 equality constraints
For improved efficiency, SDPT3 is solving the dual problem.
num. of constraints = 631
dim. of sdp var = 35, num. of sdp blk = 1
dim. of socp var = 637, num. of socp blk = 1
dim. of linear var = 595
dim. of free var = 36 *** convert ublk to lblk
SDPT3: Infeasible path-following algorithms
version predcorr gam expon scale_data
HKM 1 0.000 1 0
it pstep dstep pinfeas dinfeas gap prim-obj dual-obj cputime
0|0.000|0.000|6.3e+02|1.7e+01|5.2e+06| 1.526493e+00 0.000000e+00| 0:0:00| spchol 1 1
1|0.744|0.169|1.6e+02|1.4e+01|2.8e+06| 7.921068e+03 -8.316494e+01| 0:0:00| chol 1 1
2|0.817|0.858|2.9e+01|2.0e+00|3.8e+05| 8.862879e+03 -3.685697e+02| 0:0:00| chol 1 1
3|1.000|0.948|2.2e-05|1.1e-01|2.5e+04| 5.517002e+03 -2.310728e+02| 0:0:00| chol 1 1
4|1.000|0.581|8.8e-07|4.5e-02|1.2e+04| 2.660357e+03 -2.236909e+02| 0:0:00| chol 1 1
5|1.000|0.472|8.0e-07|2.4e-02|6.5e+03| 2.162006e+03 -2.195472e+02| 0:0:00| chol 1 1
6|0.708|0.933|2.3e-07|1.6e-03|1.5e+03| 1.073481e+03 -2.125223e+02| 0:0:00| chol 1 1
7|1.000|0.573|5.0e-09|7.1e-04|4.9e+02| 2.381452e+02 -2.124065e+02| 0:0:00| chol 1 1
8|0.971|0.603|4.6e-09|2.8e-04|3.5e+01|-1.813088e+02 -2.121119e+02| 0:0:00| chol 1 1
9|0.960|0.973|3.1e-10|8.3e-06|1.3e+00|-2.106488e+02 -2.118963e+02| 0:0:00| chol 1 1
10|0.700|0.743|1.7e-08|2.2e-06|5.4e-01|-2.110665e+02 -2.115876e+02| 0:0:00| chol 1 1
11|0.539|0.389|2.3e-08|1.3e-06|3.9e-01|-2.111280e+02 -2.114977e+02| 0:0:00| chol 2 1
12|1.000|0.167|3.5e-08|1.1e-06|2.8e-01|-2.112250e+02 -2.114725e+02| 0:0:00| chol 2 2
13|1.000|0.403|2.3e-08|6.6e-07|1.8e-01|-2.112686e+02 -2.114078e+02| 0:0:00| chol 2 2
14|0.600|0.325|1.9e-08|4.5e-07|1.4e-01|-2.113531e+02 -2.113777e+02| 0:0:00| chol 2 2
15|1.000|0.131|5.7e-07|3.9e-07|1.8e-01|-2.158019e+02 -2.113689e+02| 0:0:00| chol 5 5
16|1.000|0.015|4.7e-05|3.8e-07|3.0e+01|-1.621997e+04 -2.113685e+02| 0:0:00| chol
linsysolve: Schur complement matrix not positive definite
switch to LU factor. lu 11 30
stop: primal infeas has deteriorated too much, 2.1e+00
17|0.000|0.000|4.7e-05|3.8e-07|3.0e+01|-1.621997e+04 -2.113685e+02| 0:0:00|
number of iterations = 17
primal objective value = -2.11353103e+02
dual objective value = -2.11377667e+02
gap := trace(XZ) = 1.37e-01
relative gap = 3.23e-04
actual relative gap = 5.80e-05
rel. primal infeas (scaled problem) = 1.88e-08
rel. dual " " " = 4.45e-07
rel. primal infeas (unscaled problem) = 0.00e+00
rel. dual " " " = 0.00e+00
norm(X), norm(y), norm(Z) = 9.7e+03, 2.1e+02, 3.0e+02
norm(A), norm(b), norm(C) = 7.6e+01, 2.0e+00, 2.2e+02
Total CPU time (secs) = 0.47
CPU time per iteration = 0.03
termination code = -7
DIMACS: 1.9e-08 0.0e+00 2.7e-06 0.0e+00 5.8e-05 3.2e-04
Status: Failed
Optimal value (cvx_optval): NaN