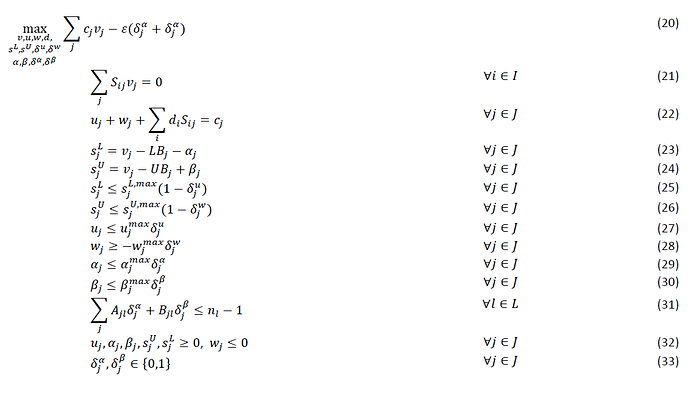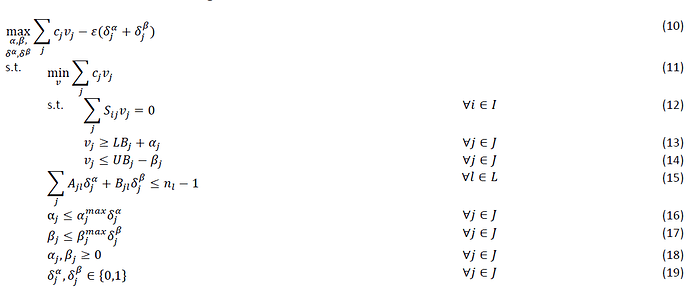Hi, I am trying to understand how to turn the following equations into matlab code that works within the CVX.
within this formulation j= set of reactions and i = set of metabolites
load ‘Ecoli_core_model.mat’
c=model.c;
S=model.S;
lb=model.lb;
ub=model.ub;
nrxns=length(model.rxns);
% FBA
cvx_begin
variables v(nrxns)
minimize (c’v)
subject to
Sv == 0
lb <= v <= ub
cvx_end
The following above is the Flux balance analysis part which would be corresponding to equation 10.
But here is about where my understanding of this stops.
And eventually we will have to try to formulate this whole thing. Which is beyond me.
This is from the journal paper “Constraint-based strain design using continuous
modifications (CosMos) of flux bounds finds new
strategies for metabolic engineering” by Cameron Cotten and Jennifer L. Reed (2013)
I’ve tried to do this but i don’t seem to be able, since I just started to learn using Matlab this February and my background is in Biochemistry, but my prof. wants me to study this but this is something I am unable to figure out on my own.
Can someone show me how to formulate this? Been reading a lot of material but I lack the understanding of some key concepts.
Best regards.
Arnar Sigurdsson